SmartANOMALY Spikefall
SmartANOMALY Spikefall
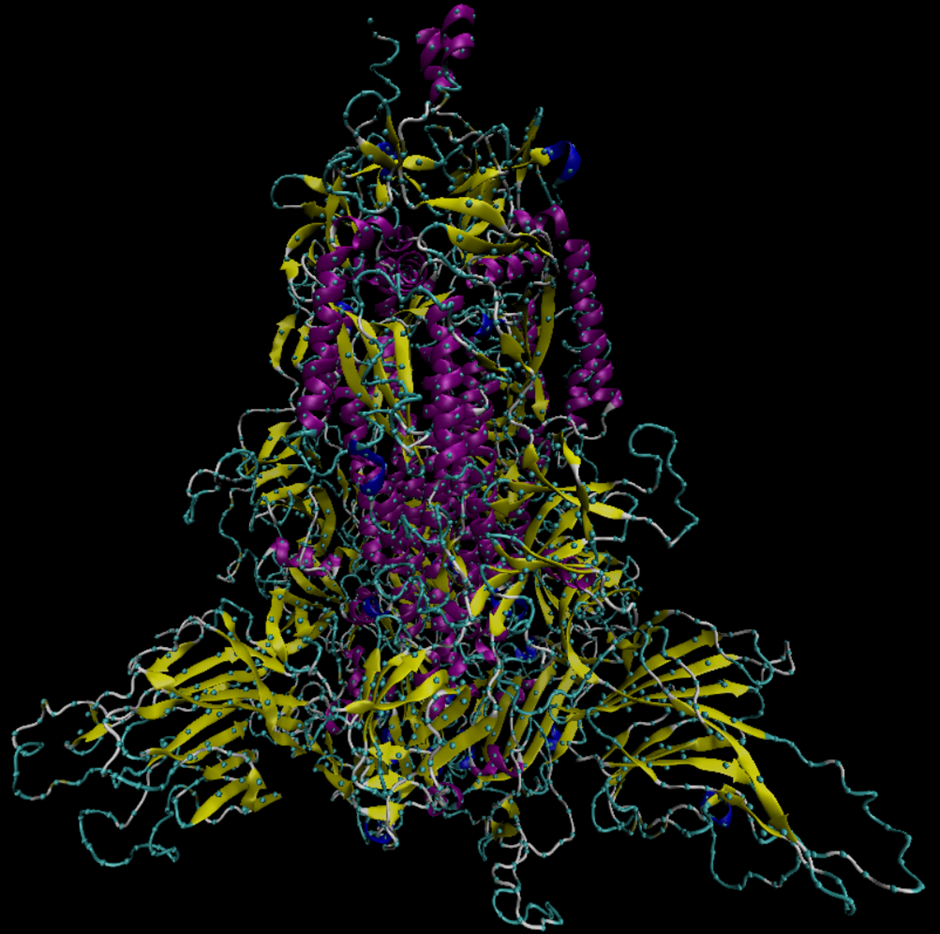
Project goal
This project was launched in the context of the fight against the COVID-19 pandemic, as a CERN openlab collaboration with the Italian Institute of Technology and CompBioMed. Through this project, we aim to propose a model based on machine learning for simulating the enhanced molecular dynamics of SARS-CoV-2’s spike glycoprotein. Proteins’ qualities can be described by their secondary structures, three-dimensional forms made of atoms groups under which protein residues (individual amino acid) can exist. The goal of SmartANOMALY Spikefall is to predict this change in structure by analysing just a few moments, or ‘frames’.
Collaborators
Project background
The energy cost for simulating proteins is high. As such, a new approach for predicting the qualities of proteins would be a very powerful tool for the research community. Such an approach could be a particular boon to the fight against the COVID-19 pandemic, with researchers seeking to accelerate their investigations.
This project is being carried out in the context of CERN's strategy for knowledge transfer to medical applications, led by CERN's Knowledge Transfer group.
Recent progress
The project started in the fourth quarter of 2020, with several approaches for classification and behaviour prediction tested immediately. Currently, we are studying the behaviour of single atoms — in particular, alpha carbons — over a short period of time. Our first observations have led us to believe that understanding the instability of an atom can provide us with insight into the long-term behaviour of a proteins’ secondary structure.
Next steps